Preparation
We start by loading the beta dispersion data generated in script 5 for the whole and core microbiomes, respectively.
data.all <- read.csv("tables/p5/boxplot_dispersion_data_whole_jjs.csv",
row.names = NULL, header = T, sep = ",")
data.all.core <- read.csv("tables/p5/boxplot_dispersion_data_core_NEW2_jjs.csv",
row.names = NULL, header = T, sep = ",")
data.all$Reef <- factor(data.all$Reef, levels = c('SCR', 'PPR', 'CCR',
'ALR', 'SIS', 'ROL',
'RNW', 'PST','PBL'))
data.all.core$Reef <- factor(data.all.core$Reef, levels = c('SCR', 'PPR',
'CCR', 'ALR',
'SIS', 'ROL',
'RNW', 'PST',
'PBL'))Then we set some plot themes.
level_order3 <- c('SCR', 'PPR', 'CCR', 'ALR', 'SIS', 'ROL', 'RNW', 'PST','PBL')
level_order <- c('Outer bay', 'Inner bay', 'Inner bay disturbed')
level_order2 <- c('Jaccard', 'Modified Gower', 'Bray Curtis', 'Unifrac',
'Generalized Unifrac', 'Weighted Unifrac')disp.pal4 <- c("royalblue4", "royalblue3", "royalblue1",
"aquamarine4", "mediumaquamarine", "aquamarine2",
"chocolate3", "chocolate2", "sienna1")
disp.pal5 <- c("midnightblue", "royalblue4", "royalblue3","darkslategrey",
"aquamarine4", "mediumaquamarine", "chocolate4", "sienna",
"chocolate3")font_theme = theme(
axis.title.x = element_text(size = 14),
axis.text.x = element_text(size = 10),
axis.title.y = element_text(size = 14),
axis.text.y = element_text(size = 8))Generate Plots
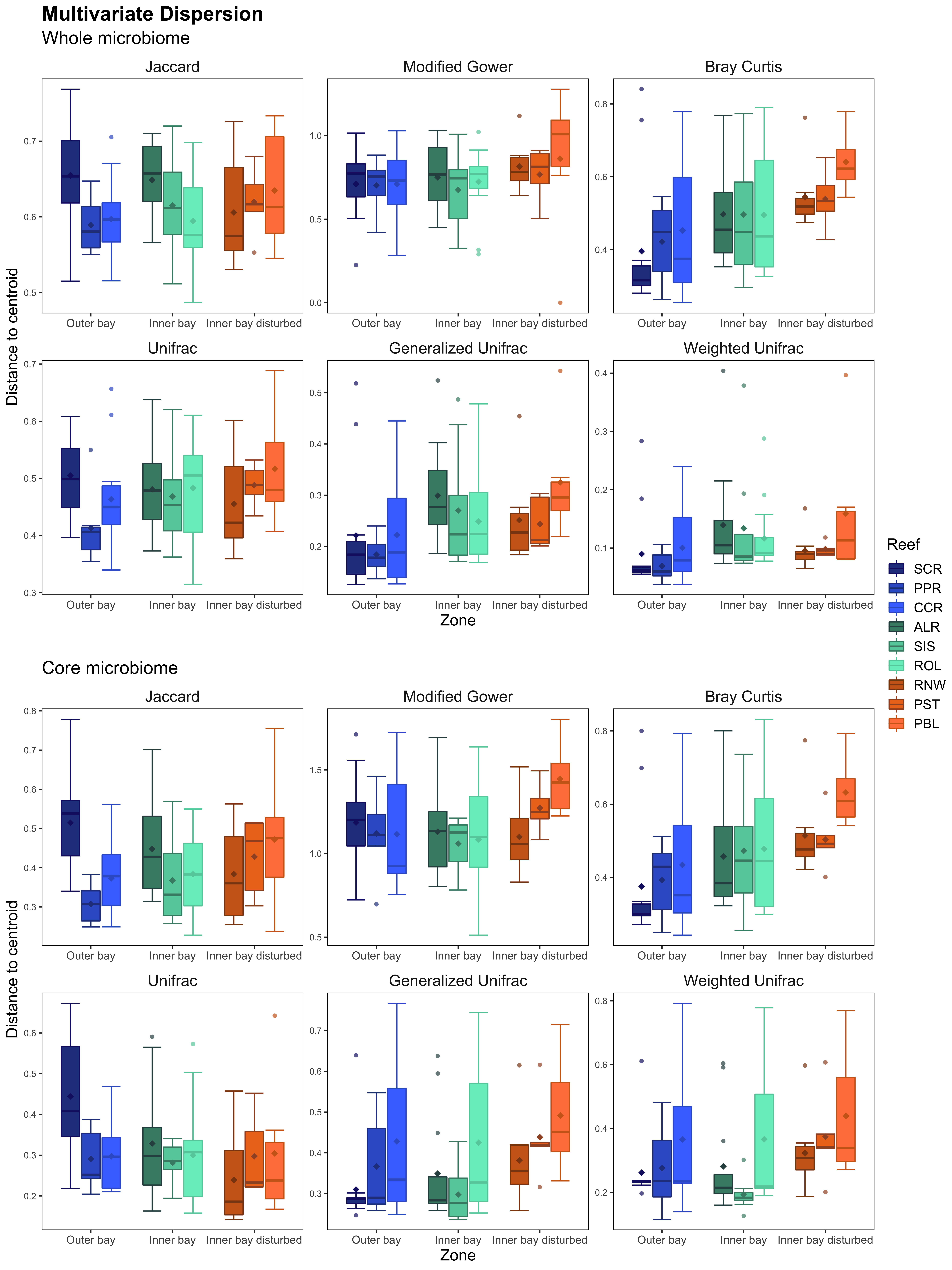
Whole community
fig4A <-
ggplot(data = data.all, aes(x = factor(Zone, level = level_order),
y=Distance_to_centroid))+
stat_boxplot(aes(colour=Reef), geom ='errorbar', alpha=1) +
geom_boxplot(aes(fill=Reef, colour=Reef), alpha=1,
outlier.alpha = 0.7, outlier.shape = 16)+
scale_colour_manual(values=disp.pal5)+
scale_fill_manual(values=disp.pal4)+
ggtitle("Multivariate Dispersion", subtitle="Whole microbiome")+
theme(plot.title=element_text(size=18, face="bold"))+
theme(plot.subtitle=element_text(size=16))+
font_theme+
stat_summary(aes(fill=Reef), fun="mean", geom="point",
size=2.5, shape=18,
position=position_dodge(width=0.75),
color=c("midnightblue", "royalblue4", "royalblue3",
"darkslategrey", "aquamarine4", "mediumaquamarine",
"chocolate4", "sienna", "chocolate3",
"midnightblue", "royalblue4", "royalblue3",
"darkslategrey", "aquamarine4", "mediumaquamarine",
"chocolate4", "sienna", "chocolate3", "midnightblue",
"royalblue4", "royalblue3","darkslategrey",
"aquamarine4", "mediumaquamarine","chocolate4",
"sienna", "chocolate3", "midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3", "midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3","midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3"), show.legend = FALSE)+
scale_x_discrete(name="Zone") +
scale_y_continuous(name="Distance to centroid")+
facet_wrap(~factor(Metric, levels=unique(Metric)),
scales="free",ncol = 3)+
theme(strip.background = element_rect(color="black", fill="white",
size=1, linetype="blank"),
strip.text.x = element_text(size = 14),
panel.border = element_rect(fill = NA, color="black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "white",colour = NA),
legend.key = element_rect(fill = "transparent", color = NA),
legend.text = element_text(size = 12),
legend.title = element_text(size = 14))
fig4ACore community
fig4B <-
ggplot(data = data.all.core, aes(x = factor(Zone, level = level_order),
y=Distance_to_centroid))+
stat_boxplot(aes(colour=Reef), geom ='errorbar', alpha=1) +
geom_boxplot(aes(fill=Reef, colour=Reef), alpha=1,
outlier.alpha = 0.7, outlier.shape = 16)+
scale_colour_manual(values=disp.pal5)+
scale_fill_manual(values=disp.pal4)+
ggtitle("", subtitle="Core microbiome")+
theme(plot.subtitle=element_text(size=16))+
font_theme+
stat_summary(aes(fill=Reef), fun="mean", geom="point", size=2.5, shape=18,
position=position_dodge(width=0.75),
color=c("midnightblue", "royalblue4", "royalblue3",
"darkslategrey", "aquamarine4", "mediumaquamarine",
"chocolate4", "sienna", "chocolate3","midnightblue",
"royalblue4", "royalblue3","darkslategrey",
"aquamarine4", "mediumaquamarine","chocolate4",
"sienna", "chocolate3","midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3","midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3", "midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna",
"chocolate3", "midnightblue", "royalblue4",
"royalblue3","darkslategrey", "aquamarine4",
"mediumaquamarine","chocolate4", "sienna", "chocolate3"),
show.legend = FALSE)+
scale_x_discrete(name="Zone") +
scale_y_continuous(name = "Distance to centroid") +
facet_wrap(~factor(Metric, levels=unique(Metric)),scales = "free", ncol = 3) +
theme(strip.background = element_rect(color="black", fill = "white",
size=1, linetype="blank"),
strip.text.x = element_text(size = 14),
panel.border = element_rect(fill = NA, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "white",colour = NA),
legend.key = element_rect(fill = "transparent", color = NA),
legend.text = element_text(size = 12),
legend.title = element_text(size = 14))
fig4BBeta Dispersion Plots Whole v Core
That’s the end of Script 6. In the next Script we use PIME as an additional tool to identify the core fish gut microbiome.
Source Code
The source code for this page can be accessed on GitHub by clicking this link.